Abstract
Deregulation of chromatin modifiers plays an essential role in the pathogenesis of medulloblastoma, the most common paediatric malignant brain tumour. Here, we identify a BMI1-dependent sensitivity to deregulation of inositol metabolism in a proportion of medulloblastoma. We demonstrate mTOR pathway activation and metabolic adaptation specifically in medulloblastoma of the molecular subgroup G4 characterised by a BMI1High;CHD7Low signature and show this can be counteracted by IP6 treatment. Finally, we demonstrate that IP6 synergises with cisplatin to enhance its cytotoxicity in vitro and extends survival in a pre-clinical BMI1High;CHD7Low xenograft model.
Similar content being viewed by others
Introduction
Medulloblastoma (MB) is the most common paediatric malignant brain tumour. Large international consortia have assembled substantial cohorts of this relatively rare tumour and have generated comprehensive genomic, epigenomic and transcriptomic datasets, which have informed a novel molecular classification of MB into four distinct molecular subgroups (WNT, SHH, G3 and G4), each of them further subdivided into subtypes with distinct prognosis and responses to therapy1,2,3,4. Despite these significant advancements in dissecting MB heterogeneity, a translational impact of these observations on how patients are treated, is still lacking. Indeed, the current standard-of-care consists of multimodal therapy, comprising surgery together with radio- and/or chemotherapy5 that does not account for the specific molecular mechanisms driving tumour growth. This regime cures a substantial proportion of patients, although it is almost invariably associated with severe side effects. Assessment of molecularly tailored therapies, including novel combinations of drugs, which take advantage of the acquired biological knowledge of MB pathogenesis, is now a priority.
Genome-wide sequencing6,7,8,9 has identified mutations in genes regulating essential epigenetic mechanisms, such as histone demethylases, deacetylases, lysine methyltransferases and chromatin remodellers, as well as overexpression of Polycomb-group (PcG) proteins10,11,12, particularly in G4 MB, the least understood of all subgroups, despite being the most common and associated with poor prognosis. The PcG protein BMI1, a key regulator of neural stem cell (NSC) self-renewal13,14, is upregulated in a broad range of cancers, including brain tumours such as glioma15, where it correlates with advanced clinical stage and poor prognosis16. In MB, the highest expression level of BMI1 is associated with the G4 subgroup where it sustains tumour growth10,17. We previously demonstrated that high expression of BMI1 together with loss-of-function of the ATP-dependent chromatin remodeller Chromodomain-Helicase-DNA-binding protein 7 (CHD7) cooperates in inducing MB formation when glutamatergic progenitor cells, expressing the proneural transcription factor Math1, are targeted in a sleeping beauty-driven forward genetic screening in the mouse10. Moreover, we showed that a BMI1High;CHD7Low signature is found in a proportion of G4 MB, a subgroup recently shown to have an ontogenetic link to unipolar brush cell (UBC) progenitor cells, which develop from a Math1+ progenitor18.
Epigenetic mechanisms sustain cancer growth not only through regulation of the expression of genes controlling cell proliferation but also through modulation of pathways involved in cellular metabolism19. Energy metabolism is altered in cancer cells, which display enhanced glycolysis favouring production of lactate from glucose even in the presence of oxygen20 with other metabolic processes, including protein, nucleic acid and lipid biosynthesis, also being enhanced as part of the tumour metabolic reprogramming. Chromatin remodelling via histone modifications and DNA hypermethylation contributes to repression of gluconeogenesis enzymes and consequent activation of glycolysis in different cancers21,22,23. Aerobic glycolysis provides up to 60% of ATP production in MB24, but whether it is regulated by epigenetic mechanisms in this tumour type is currently unknown.
Energy production is finely tuned to cellular function and state, and is known to be modulated by several signalling pathways, such as Akt/mTOR, in which phosphoinositides and inositol pyrophosphates are key players25,26. Inositols (phosphorylated forms of myoinositol and inositol hexakisphosphate, IP6) are highly energetic polyphosphate molecules that play a critical role in a wide variety of physiological and pathological properties of the cell, possibly through phosphorylation-dependent modulation of multiple signalling pathways27. Deregulated metabolism of inositol has been described in different diseases, including cancer where it exerts an antitumour activity mainly modulating cell growth and inducing differentiation and apoptosis28,29,30. However, whether inositol can have therapeutic efficacy and a role in sustaining MB growth has not yet been elucidated.
Here, we perform epigenomic, transcriptomic and proteomic characterisation of patient-derived MB cells modelling the BMI1High;CHD7Low molecular convergence, as well as generate genetically engineered and xenograft models to elucidate the molecular mechanisms underpinning BMI1 and CHD7 convergence, its impact on MB metabolism and its druggability at preclinical level.
Results
Modelling a BMI1High;CHD7Low signature in mice leads to increased number of EOMES+ UBC, but no medulloblastoma
To test the oncogenic potential of the BMI1High;CHD7Low molecular signature previously identified10, we generated mice overexpressing Bmi1 in Math1+ progenitors (Math1Cre;STOPFloxBmi1), which were then further crossed with Chd7 floxed mice (Chd7flox) to obtain Math1Cre;STOPFloxBmi1;Chd7f/+. Compound mice were healthy and did not develop medulloblastoma during observation of up to 12 months (0/13mice). To assess whether developmental abnormalities were elicited by these genetic modifications, we focussed on neuronal populations originating from Math1+ progenitors in the cerebellum. Math1 is expressed in a common progenitor giving rise to both Unipolar Brush Cells (UBC) and Granule Cells (GC)12,18,31; therefore we analysed the number of UBC and GC in Math1Cre;STOPFloxBmi1;Chd7f/+ compared to single transgenic controls, as assessed by immunostaining for EOMES (Eomesodermin) and NeuN, respectively. NeuN is not expressed in UBC32. We observed an increased number of EOMES-expressing UBC in the cerebellum of the compound mutants at both P7 and P21 stages (Fig. 1a, b and Supplementary Fig. S1a–c), whilst the number of mature GC in the cerebellar cortex was reduced (Fig. 1c and Supplementary Fig. S1d). Importantly, no significant changes were observed in the proliferation potential of Granule Cell Progenitors (GCP) at the clonal expansion stage (P7) in these mice (Supplementary Fig. S1e, f).
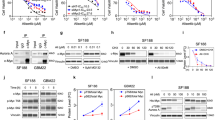
a, b Colours identify STOPFloxBmi1 (yellow), Chd7f/+ (orange), Math1Cre;STOPFloxBmi1 (green) or Math1Cre;STOPFloxBmi1;Chd7f/+ (pink) genotypes throughout the figure. EOMES IHC staining and quantification of UBC in the cerebellum at P7 (a) or P21 (b) developmental stages. a n = 2 biological independent animals per STOPFloxBmi1 and Math1Cre;STOPFloxBmi1 genotypes, n = 3 biological independent animals per Chd7f/+ and Math1Cre;STOPFloxBmi1;Chd7f/+ genotypes. b n = 3 biological independent animals per STOPFloxBmi1 and Chd7f/+ genotypes, n = 4 biological independent animals per Math1Cre;STOPFloxBmi1;Chd7f/+ genotype. One-way ANOVA. c NeuN IHC staining and quantification of GC in the cerebellum at P7. n = 2 biological independent animals per STOPFloxBmi1 and Math1Cre;STOPFloxBmi1 genotypes, n = 3 biological independent animals per Chd7f/+genotype, n = 4 biological independent animals per Math1Cre;STOPFloxBmi1;Chd7f/+ genotype, one-way ANOVA. d Heatmap showing z-scores of UBC marker expression in the human upper rhombic lip (light blue), cerebellum (yellow) and cerebellar cortex (green) from 8 to 17 pcw. e Heatmap showing relative expression of BMI1 and CHD7 in a collection of hNCS lines isolated from the cerebellum (yellow) or striatum (green) as compared to MB cell lines (pink). f qPCR analysis of EOMES expression level in hNSC upon CHD7 silencing (BMI1High;CHD7Low, purple) compared to control (CTRL, violet). n = 4 biologically independent experiments, two-tailed unpaired t test. g Cell proliferation assay of CDH7-silenced or control cells. n = 6 biologically independent experiments, two-way ANOVA. All graphs report mean ± SEM. P values: *P < 0.05, **P < 0.01 or ****P < 0.0001. Scale bars = 100 µm. Source data are provided as a Source Data file.
Next, we interrogated the Human Allen Brain Atlas to map the expression pattern of all genes reported to be expressed, although not necessarily exclusively, in UBC18 during the development of the human cerebellum. We found increased expression of all genes, including the UBC-specific marker EOMES, from the 13th gestational week onwards (Fig. 1d). Expression levels for BMI1 and CHD7 were also investigated in a collection of human neural stem cell (hNSC) lines derived from foetal cerebellum at various stages of development (Glioma Cellular Genetics Resource) (Fig. 1e). The hNSC line derived from a 13-week sample had the highest expression of BMI1 and CHD7 and this time point was therefore chosen to examine the effect of silencing CHD7 expression, using shRNAs (Supplementary Fig. S1g), on UBC marker expression. We found increased expression of EOMES (Fig. 1f) in hNSC upon CHD7 silencing in a BMI1-expressing context, which led to increased proliferation of the cells (Fig. 1g), but no MB formation upon orthotopic injection into the neonatal cerebellum (0/10 mice at 8.5 months).
We show that a BMI1High;CHD7Low signature leads to an increased number of UBC and reduced number of mature GC in the developing cerebellum, raising the possibility that it favours the differentiation of a common Math1+ progenitor towards a UBC lineage. UBC is the most promising putative cell of origin of G4 MB; hence, our results represent a strong indicator of a potential role for BMI1 and CHD7 in the pathogenesis of this MB subgroup, although they are not sufficient on their own to elicit MB formation in either genetically engineered mice or hNSC xenografted models.
mTOR signalling is activated in G4 MB lines, but not cerebellar hNSC, with a BMI1High;CHD7Low signature
To gain further insights into the differences between the role played by the BMI1;CHD7 molecular convergence in a neoplastic and non-neoplastic context, we performed an integrated analysis of the methylome and transcriptome of ICb1299 and CHLA-01-Med MB cell lines. These cell models were chosen because no stable patient-derived xenografts (PDX) have been obtained from BMI1High;CHD7Low G4 patients, rendering in vitro genetic modification of BMI1-overexpressing cell lines the only suitable model to assess the contribution of the signature to MB pathogenesis. ICb1299 and CHLA-01-Med MB cell lines display a G3/G4 subgroup affiliation10,31,33,34,35, reflecting a widely recognised boundary of plasticity between these two subgroups31,36. Modelling of the signature was achieved in these lines by CHD7 silencing in the context of BMI1 overexpression as compared to normal cerebellum as previously described10. Integration of the datasets of the cell model with those obtained in G4 MB tumour samples4 ensured that the focus would be on events shared between these cell models and subgroup-specific tissue samples (Fig. 2a). We compared samples with and without the signature and identified 214 differentially expressed genes (DEG) with 361 differentially methylated probes (DMP) in MB cells and 2454 DEG with 138 DMP in G4 MB tissues (Supplementary Fig. S2a). Canonical pathway analysis identified significantly enriched pathways related to both signalling and metabolism, including lipids and phospholipid metabolism and energy production, in each of the four datasets (Fig. 2b and Supplementary Fig. S2b). Comparative analysis of the results revealed a single common and conserved pathway Superpathway of Inositol Phosphate Compounds (Fig. 2c), which was not identified when significant DEG between G3 MB tumours with and without BMI1High;CHD7Low signature was analysed (Supplementary Fig. S2c), in keeping with the correlation between the signature and this metabolic pathway being specifically pertinent to the G4 subgroup.
a Schematic representation of the comparative analysis between BMI1High;CHD7Low MB cell lines and G4 patients with and without the signature performed to identify genes differentially expressed (DE) and differentially methylated probes (DMP) and commonly deregulated pathways. b Pie charts representing canonical pathways differentially enriched between MB cells or patients with or without BMI1High;CHD7Low signature identified in RNA-Seq or DNA methylation analysis. Canonical pathways are classified based on IPA categories list and colour-coded accordingly. c Venn diagram of canonical pathways enriched for genes differentially expressed (blue and light-green diagrams as coloured in a and b) or methylated (olive green and red as coloured in a and b) in BMI1High;CHD7Low cell lines and GR4 MBs with the matching signature. Highlighted the superpathway of phosphoinositol compounds (phosphoinositols), which is common to all the datasets analysed. d Bubble plot showing significant Reactome pathways obtained in phosphoproteomic analysis of BMI1High;CHD7Low MB lines. Bubbles are coloured based on FDR values, and size is proportional to the number of genes of specific pathways. e, f Cell viability assays of MB cells (e) or hNSC (f) with (BMI1High;CHD7Low) or without the signature (CTRL) upon 72 h of treatment with increasing concentrations of FLT3 inhibitor (TCS). Histograms represent the percentages of viable cells relative to non-treated cells (top). Measurement of Area Under Curve (AUC) (bottom) to compare the overall response to treatment. e n = 6 biologically independent experiments, (f) n = 4 biologically independent experiments, two-way ANOVA. g, h Cell viability assays of MB cells (g) or hNSC (h) with (BMI1High;CHD7Low) or without the signature (CTRL) upon 72 h of treatment with increasing concentrations of MET inhibitor (PHA). Histograms represent the percentages of viable cells relative to non-treated cells (top). Measurement of the area under the curve (AUC) (bottom) to compare the overall response to treatment. g n = 6 biologically independent experiments, h n = 4 biologically independent experiments, two-way ANOVA. i Western blot and quantification of phosphorylated/total RPS6 (pRPS6, Ser240/244) in MB cells. GAPDH immunoreactivity was used to normalise protein loading. n = 4 biologically independent experiments, one-way ANOVA. j Cell viability assays of MB cells with (BMI1High;CHD7Low, red) or without the signature (CTRL, green) upon 72 h of treatment with increasing concentrations of mTOR pathway inhibitors (rapamycin and torin). Histograms represent the percentages of viable cells relative to non-treated cells (top). Measurement of Area Under Curve (AUC) (bottom) to compare the overall response to treatment. n = 5 biologically independent experiments, two-way ANOVA. k Western blot and quantification of phosphorylated/total RPS6 (pRPS6, Ser240/244) after 24 h of treatment with increasing concentrations of IP6 in control (green) or BMI1High;CHD7Low (red) MB cells. GAPDH immunoreactivity was used to normalise protein loading. n = 4 biologically independent experiments, two-way ANOVA. All graphs report mean ± SEM. P values: *P < 0.05, **P < 0.01, ***P < 0.001 or ****P < 0.0001. Source data are provided as a Source Data file.
Phosphorylated forms of myoinositol and inositol hexakisphosphate (IP6) are highly energetic polyphosphate molecules regulating many physiological cell properties through modulation of phosphorylation-dependent pathways27. To investigate phosphorylation-dependent events, possibly related to the inositol metabolism, we performed a phosphoproteomic analysis of BMI1High;CHD7Low MB lines and controls. We identified 1499 significantly deregulated phosphopeptides, 684 hyper- and 815 hypophosphorylated (Supplementary Fig. S2d), and interrogation of this dataset with Reactome pathway analysis confirmed modulation of phosphoinositol metabolism (Fig. 2d). Moreover, we found an enrichment in signalling mediated by receptor tyrosine-kinase (RTK) and several related pathways, including MAPK/ERK, as previously described10. Because this pathway is deregulated also in hNSC10, we reasoned that it must not be sufficient to trigger MB formation. MET and FLT3 RTKs are also enriched for in our phosphoproteomic analysis (Fig. 2d) with the expression of both receptors being significantly higher in BMI1High;CHD7Low MB than in controls, whilst no significant difference was observed in hNSC (Supplementary Fig. S2e, f). To assess the functional impact of the observed MB-specific increased expression of these two RTKs, we analysed cell viability of MB or hNSC cells upon treatment with TCS359 and PHA665752, specific inhibitors of FLT3 and MET, respectively. Pharmacologic inhibition of FLT3 impaired cell viability specifically in MB modelling the signature (Fig. 2e), without affecting hNSC (Fig. 2f). Moreover, BMI1High;CHD7Low MB cells were more susceptible to MET inhibition (Fig. 2g), although hNSC was also affected, albeit in the opposite direction, i.e. those modelling the signature were more resistant (Fig. 2h). These results validate the in silico prediction of enhanced signalling through the tyrosine-kinase receptors FLT3 and MET in BMI1High;CHD7Low MB but not in hNSC with the same molecular signature.
RTKs modulate a variety of downstream phosphorylation-dependent signalling cascades37,38. Integrated pathway analysis of DE genes and phosphorylation-dependent proteins predicted an impact on mTOR signalling in BMI1High;CHD7Low MB, a modulation that is also observed in tumour samples with the same signature (Fig. 2d and Supplementary Fig. S2h). To validate this in silico prediction, we analysed phosphorylation of ribosomal protein S6 (RPS6), a well-characterised target of the mTOR pathway. We confirmed the overactivation of the mTOR pathway in MB cells modelling the signature, which is dependent on both CHD7 silencing and concomitant expression of BMI1 (Fig. 2i and Supplementary Fig. S2f, g). FLT3 and MET activate AKT/mTOR signalling39,40,41. In line with the observation that BMI1High;CHD7Low MB cells show an increased dependence on FLT3 and MET signalling, we found that their inhibition decreases mTOR activation specifically in BMI1High;CHD7Low MB cells (Supplementary Fig. S2i, j). Importantly, no impact on RPS6 phosphorylation was observed in BMI1High;CHD7Low hNSC (Supplementary Fig. S2k), raising the possibility that the impact on the mTOR pathway is MB-specific. Impaired cell viability was observed upon treatment with two mTOR inhibitors (rapamycin and torin) in BMI1High;CHD7Low MB as compared to control (Fig. 2j and Supplementary Fig. S2i) and IP6 treatment reduced mTOR activation specifically in BMI1High;CHD7Low MB cells as compared to cells without the signature (Fig. 2k).
Our data show deregulation of inositol metabolism and RTKs in MB cells and G4 tumour samples with a BMI1High;CHD7Low signature and activation of the mTOR signalling pathway in these cells, but not in hNSC with the same molecular signature.
A BMI1High;CHD7Low signature induces metabolic adaptation in MB but not in hNSC
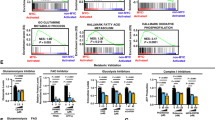
Both phosphoinositols and mTOR pathways regulate cell viability primarily via modulation of energy production42,43,44,45. We, therefore, studied the energetic state of BMI1High;CHD7Low MB cells and analysed mitochondrial respiration and glycolysis, by measuring oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), respectively. OCR was decreased in BMI1High;CHD7Low MB cells with a reduction in basal, ATP-linked and maximal respiration, indicating an impaired mitochondrial function (Fig. 3a and Supplementary Fig. S3a) rather than a reduced number of mitochondria, as no significant changes in mtDNA were detected (Supplementary Fig. S3b). Consistently, staining with a potential-dependent MitoTracker dye showed increased mitochondrial membrane potential, suggestive of mitochondrial hyperpolarisation in BMI1High;CHD7Low MB cells (Supplementary Fig. S3c). Intriguingly, mitochondrial dysfunction is among the significantly enriched pathways found in BMI1High;CHD7Low G4 MB tissue samples (Supplementary Fig. S2b). In contrast, increased ECAR with enhanced glycolysis and glycolytic capacity was detected in MB cells with the signature (Fig. 3b). These results indicate metabolic adaptation in BMI1High;CHD7Low MB cells, leading to impaired mitochondrial respiration and enhanced aerobic glycolysis.
a Analysis of oxidative consumption rate (OCR) using seahorse extracellular flux assay. BMI1High;CHD7Low (red) or control (green) MB cells were sequentially treated with oligomycin, P-trifluoromethoxy carbonyl cyanide phenylhydrazone (FCCP) or a combination of antimycin A and rotenone at the indicated time points (dashed lines). Histograms (right) show OCR production during basal, ATP-linked and maximal respiration. n = 4 biologically independent experiments for CTRL or n = 5 biologically independent experiments for BMI1High;CHD7Low, two-tailed unpaired t test. b Analysis of extracellular acidification rate (ECAR) using seahorse extracellular flux assay. BMI1High;CHD7Low or control MB cells were sequentially treated with glucose, oligomycin or 2-deoxy-d-glucose (2DG) at the indicated time points (dashed lines). Histograms (right) show ECAR representing glycolysis and glycolytic capacity. n = 7 biologically independent experiments, two-tailed unpaired t test. c Quantification of ATP content in BMI1High;CHD7Low or control MB cells, normalised on quantity of total protein. n = 9 biologically independent experiments, two-tailed unpaired t test. d Quantification of ADP/ATP ratio in BMI1High;CHD7Low or control MB cells. n = 10, two-tailed unpaired t test. e Analysis of ECAR using seahorse extracellular flux assay. BMI1High;CHD7Low or control MB cells were incubated for 24 h with IP6 1 mM and then sequentially treated with glucose, oligomycin or 2-deoxy-d-glucose (2DG) at the indicated time points (dashed lines). Histograms (right) show ECAR representing glycolysis and glycolytic capacity. n = 4 biologically independent experiments, one-way ANOVA. f Analysis of oxidative consumption rate (OCR) using seahorse extracellular flux assay. BMI1High;CHD7Low (purple) or control (violet) hNSC cells were sequentially treated with oligomycin, FCCP or a combination of antimycin A and rotenone at the indicated time points (dashed lines). Histograms (right) show OCR production during basal, ATP-linked and maximal respiration. n = 3 biologically independent experiments, two-tailed unpaired t test. g Analysis of extracellular acidification rate (ECAR) using seahorse extracellular flux assay. BMI1High;CHD7Low or control hNSC cells were sequentially treated with glucose, oligomycin or 2-deoxy-d-glucose (2DG) at the indicated time points (dashed lines). Histograms (right) show ECAR representing glycolysis and glycolytic capacity. n = 3 biologically independent experiments, two-tailed unpaired t test. h Quantification of ATP content in BMI1High;CHD7Low or control hNSC cells, normalised on the quantity of total protein. n = 9 biologically independent experiments, two-tailed unpaired t test. i Quantification of ADP/ATP ratio in BMI1High;CHD7Low or control hNSC cells. n = 10 biologically independent experiments, two-tailed unpaired t test. All graphs report mean ± SEM. P values: *P < 0.05, **P < 0.01, ***P < 0.001 or ****P < 0.0001. Source data are provided as a Source Data file.
To assess whether this finding could be useful for future use in the clinic, we identified BMI1High;CHD7Low G4 MB in a cohort of patients where magnetic resonance spectroscopy (MRS) analysis had been performed on surgically resected tumour samples46. We show an increased quantity of leucine and valine, two branched-chain amino acids (BCAA), in MB with the signature as compared to samples without the signature, in keeping with an enhanced glycolytic status also in the tumour tissue (Supplementary Fig. S3d). Consistently, we found increased expression of HK2, PFKP, ENO4, PDK1 and LDHB, encoding key enzymes regulating glucose metabolism, in BMI1High;CHD7Low G4 MB tumour samples (Supplementary Fig. S3e) but not in G3 tumours with the same signature (Supplementary Fig. S3f), further confirming the relevance of the BMI1High;CHD7Low signature specifically for G4 MB subgroup.
Mitochondria and glycolysis produce ATP; hence, we determined the total quantity of ATP produced in a steady state and found that BMI1High;CHD7Low MB cells produce significantly more ATP than controls (Fig. 3c). Moreover, the analysis of ADP/ATP ratio showed no significant difference (Fig. 3d), indicating that ATP utilisation is not different between MB cells with or without the signature and suggesting that the observed deregulation of OCR and ECAR ultimately impacted energy production. Importantly, IP6 treatment reverted the metabolic adaptation observed in BMI1High;CHD7Low MB cells by decreasing ECAR and glycolytic capacity (Fig. 3e), without affecting OCR (Supplementary Fig. S3g). In contrast, BMI1High;CHD7Low hNSC showed increased mitochondrial (Fig. 3f and Supplementary Fig. S3h, i) and glycolytic functions (Fig. 3g) compared to isogenic hNSC without the signature. However, BMI1High;CHD7Low hNSC show a reduced total ATP production (Fig. 3h) possibly due to increased ATP utilisation as suggested by increased ADP/ATP ratio compared to hNSC without the signature (Fig. 3i). Interestingly, IP6 treatment does not affect ECAR, glycolysis and glycolytic capacity of BMI1High;CHD7Low hNSC (Supplementary Fig. S3j), indicating that IP6-mediated inhibition of glycolytic function is achieved specifically in MB context.
Taken together, our data show that a BMI1High;CHD7Low signature induces a metabolic adaptation in G4 MB, but not in hNSC with the same signature, and that it can be counteracted by IP6 treatment.
IP6 sensitivity in BMI1High;CHD7Low MB cells is BMI1-dependent
Taking into account that inositol pathway dysregulation, through IP6 treatment, inhibits mTOR pathway activation and the metabolic adaptation mediated by the signature in G4 MB cells, we set out to assess their impact on tumour cell viability. We found impaired cell proliferation in BMI1High;CHD7Low MB lines at all tested IP6 concentrations compared to control MB cells without the signature, which were only affected by the highest concentration used (Fig. 4a, b and Supplementary Fig. S4a). No induction of apoptosis was found upon treatment (Supplementary Fig. S4b), suggesting a cytostatic rather than a cytotoxic effect mediated by IP6. In contrast, treatment of hNSC with or without the signature did not affect cell viability (Fig. 4c), raising the possibility that IP6 could be a novel signature-specific treatment for MB with a very low toxicity on the surrounding developing cerebellum.
a Cell proliferation assays of control (CTRL, green), BMI1High;CHD7Low (red) or with concomitant BMI1 silencing (BMI1High;CHD7Low + shBMI1, blue) MB cells upon 72 h and treated with increasing concentrations of IP6. n = 6 biologically independent experiments, two-way ANOVA used to calculate p-values (colour-coded) relative to not treated cells. b Cell viability assays of control, BMI1High;CHD7Low or with concomitant BMI1 silencing MB cells upon 24 h of treatment with increasing concentrations of IP6. Measurement of the area under the curve (AUC) to compare the overall response to treatment. n = 6 biologically independent experiments, one-way ANOVA. c Cell viability assays of control (violet) or BMI1High;CHD7Low (purple) hNSC upon 24 h of treatment with increasing concentrations of IP6. Measurement of the area under the curve (AUC) to compare the overall response to treatment. n = 6 biologically independent experiments, two-tailed unpaired t test. d Distribution of genomic annotations and heatmaps of BMI1 ChIP-Seq peak enrichments in control (CTRL) or BMI1High;CHD7Low MB cells. Heatmaps are centred at TSS and ordered by BMI1 intensity. Signals were clustered based on the distribution of BMI1 surrounding the promoters. One-sided χ2 test P value and effect size (E.s.) are reported. e Heatmaps of BMI1 and H3K27me3 ChIP-Seq peak enrichment on BMI1-bound promoters in CTRL or BMI1High;CHD7Low MB cells (left panels). Heatmaps are centred at TSS and ordered by BMI1 intensity. Signals were clustered based on the distribution of BMI1 and H3K27me3 surrounding the promoters. Venn diagrams showing the overlap between genes with BMI1 and H3K27me3 on their promoter (middle panels). Bubble plots showing GO biological processes significantly enriched for genes with promoter co-bound by BMI1 and H3K27me3 in control (green) or BMI1High;CHD7Low (red) MB cells (right panels). Bubbles are coloured based on FDR values and size is proportional to the number of genes of specific GO terms. f Venn diagrams showing the overlap between genes with BMI1, H3K27me3 and H3K4me3 on their promoter (top panels). Bubble plots showing GO biological processes significantly enriched for genes with promoter co-bound by BMI1, H3K27me3 and H3K4me3 in control (green) or BMI1High;CHD7Low (red) G4 MB cells (bottom panels). Bubbles are coloured based on FDR values and size is proportional to the number of genes of specific GO terms. g Venn diagram showing the overlap between a gene involved in phosphoinositol compound metabolism and gene with BMI1, H3K27me3 and H3K4me3 on their promoter (top) in BMI1High;CHD7Low MB cells. Western blot and quantification of PPIP5K2 expression after 24 h of treatment with increasing concentrations of IP6 in control (green) or BMI1High;CHD7Low (red) MB cells. GAPDH immunoreactivity was used to normalise protein loading. n = 3 biologically independent experiments, two-way ANOVA. h Heatmap showing inositol-related pathways significantly enriched for genes differentially methylated in BMI1High;CHD7Low (red) or BMI1High;CHD7Low with concomitant BMI1 silencing (blue). All graphs report mean ± SEM. P values: *P < 0.05, **P < 0.01 or ****P < 0.0001. Source data are provided as a Source Data file.
A PRC (Polycomb Repressive Complex)-dependent CHD7-mediated epigenetic regulation of lipid inositol signalling in BMI1High;CHD7Low MB cells
Because BMI1 expression mediated IP6 sensitivity in a context of low CHD7 expression (Fig. 4a, b and Supplementary Fig. S4a), we set out to clarify the signature-specific role of BMI1 in the inositol- related response. We interrogated the genome-wide distribution of BMI1 in the two MB cell lines using chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq) and focussed on the shared peaks. We found a significant redistribution of BMI1 binding across the genome in a CHD7Low cellular context, with a two-fold enrichment in the percentage of promoter occupancy (Fig. 4d). Next, we assessed differential BMI1 binding at the promoter of genes related to phosphoinositol metabolism upon CHD7 silencing and identified FLT3, MET and phosphatidylinositol-4-phosphate 5-kinase type 1 beta (PIP5K1B) uniquely bound by BMI1 in control cells with the expression of FLT3 and MET increasing upon BMI1 silencing (Supplementary Fig. S4c), suggesting a direct regulation via BMI1. To link the observation of BMI1 binding to a PRC (Polycomb Repressive Complex)-mediated epigenetic regulation, we identified those promoters that were also marked by histone H3 lysine 27 trimethylation (H3K27me3) and found a significant overlap in MB cells with and without signature (210 and 475 representing 76.1 and 66.4% of all BMI1 peaks on promoters, respectively, Fisher’s exact test: p < 0.0001) (Fig. 4e and Supplementary Fig. S4d). Gene Ontology (GO) enrichment analysis of the genes with BMI1 and H3K27me3 promoter co-occupancy identified activation of phospholipase C (PLC) activity specifically in cells with the signature (Fig. 4e and Supplementary Fig. S4e). PLC produces diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3) and consequently regulates inositol lipid signalling pathways47,48. Thus, our data suggest a BMI1-dependent CHD7-mediated impact on inositol signalling in MB cells.
A BMI1-mediated dynamic epigenetic regulation of inositol lipid signalling through poised promoters in BMI1High;CHD7Low MB cells
Because chromatin remodelling factors can dynamically regulate gene expression through bivalent domains on promoters marked by both the repressive H3K27me3 and the activating trimethylation on lysine 4 of histone H3 (H3K4me3) marks49, we sought to identify genes with BMI1-bound promoter regions also harbouring H3K27me3 and H3K4me3 marks and found 149 and 64 genes representing 20.8% or 23.2% of all BMI1-bound promoters in MB cells without or with BMI1High;CHD7Low signature. As expected, GO enrichment analysis revealed a shared enrichment for biological processes related to nervous system development and neuron fate commitment (Fig. 4f and Supplementary Fig. S4f), in line with the observations of bivalent domains mainly marking key developmental genes49. Intriguingly, we also found activation of PLC activity enriched for in BMI1High;CHD7Low MB (Fig. 4f and Supplementary Fig. S4f), suggesting a BMI1-mediated dynamic epigenetic regulation of inositol lipid signalling through poised promoters. Finally, we assessed whether any of the 64 genes with the promoter co-occupied by BMI1, H3K27me3 and H3K4me3 were related to the Superpathway of Phosphoinositol Compounds. PPIP5K2, an inositol pyrophosphate kinase that synthesises 1-IP7 from IP650, was identified specifically in BMI1High;CHD7Low MB cells, with the H3K27me3 mark being lost at its promoter upon BMI1 silencing (Fig. 4g and Supplementary Fig. S4g). Because PPIP5K2 expression was downregulated upon IP6 administration specifically in BMI1High;CHD7Low MB cells (Fig. 4g), our data raise the possibility that epigenetic regulation of PPIP5K2 regulates inositol metabolism and enhances IP6-induced cytotoxicity in these cells. Moreover, inositol-related canonical pathways enriched for genes differentially methylated upon CHD7 silencing were found to switch from a hyper- to hypomethylated status (Fig. 4h) upon concomitant BMI1 silencing, including PPIP5K2 (Supplementary Fig. S4h), providing additional support to the conclusion that the observed BMI1-dependent deregulation of inositol metabolism is epigenetically regulated (Supplementary Fig. S4i).
In conclusion, our data show that the BMI1High;CHD7Low specific response to inositol pathway alteration, through IP6 treatment, is dependent on BMI1-mediated epigenetic regulation.
IP6 synergises with cisplatin to reduce cell viability in BMI1High;CHD7Low MB cells
Cisplatin and etoposide are chemotherapeutic agents currently used in the treatment of MB patients5. To begin assessing the potential value of inositol treatment as adjuvant therapy, we treated BMI1High;CHD7Low MB cells with either of the two chemotherapeutic agents alone or in combination with IP6. We found that control cells are resistant to the range of cisplatin concentration used in vitro, while treatment with etoposide caused a decreased cell viability (Fig. 5a). In contrast, BMI1High;CHD7Low cells are sensitive to all the tested doses of cisplatin, while etoposide only induces decreased cell viability at the highest concentration (Fig. 5a). Combination treatments with IP6 showed that it synergises with cisplatin, but not etoposide, enhancing its cytotoxic activity specifically in BMI1High;CHD7Low cells (Fig. 5b, c).
a Cell viability assays of MB cells without (CTRL, shades of green, left panels) or with (BMI1High;CHD7Low, shades of red, right panels) the signature upon 72 h of treatment with increasing concentrations of cisplatin (CISP) or etoposide (ETOP). Histograms represent the percentages of viable cells relative to non-treated cells. n = 6 biologically independent experiments, one-way ANOVA. b Cell viability assays of control (left) or BMI1High;CHD7Low (middle) MB cells upon 72 h of combination treatment with increasing concentrations of cisplatin and IP6. Histograms represent the percentages of viable cells relative to non-treated cells. n = 6 biologically independent experiments, two-way ANOVA. Surface plots (right panels) of synergy scores obtained after combination treatment of IP6 and cisplatin. n = 3 biologically independent experiments. c Cell viability assays of control (left) or BMI1High;CHD7Low (middle) MB cells upon 72 h of combination treatment with increasing concentrations of etoposide and IP6. Histograms represent the percentages of viable cells relative to non-treated cells. n = 6 biologically independent experiments, two-way ANOVA. Surface plots (right panels) of synergy scores obtained after combination treatment of IP6 and etoposide. n = 3 biologically independent experiments. d Experimental design to assess the combinatorial effect of IP6 and cisplatin treatment administered in drinking water or through intraperitoneal injection (i.p.), respectively. e, f Kaplan–Meier survival curves of mice orthotopically xenografted with BMI1High;CHD7Low (e) or control (f) MB cells treated with vehicle, IP6, cisplatin or both (IP6 + cisplatin). n = 6 biological independent animals per cisplatin-treated group, n = 7 biological independent animals per IP6 + cisplatin-treated groups, n = 8 biological independent animals per CTRL IP6-treated group, n = 9 biological independent animals per BMI1High;CHD7Low IP6-treated group, n = 7 biological independent animals per CTRL vehicle-treated group, n = 9 biological independent animals per BMI1High;CHD7Low vehicle-treated group, two-tailed P values determined by log-rank test. g Quantification of the area of the tumours developed in mice treated as described in (e) and (f). n = 8 biological independent animals per BMI1High;CHD7Low vehicle-treated group, n = 6 biological independent animals per BMI1High;CHD7Low IP6 + Cisp-treated group, n = 2 biological independent animals per CTRL vehicle-treated group, n = 4 biological independent animals per CTRL IP6 + Cisp-treated group, one-way ANOVA. h, i Quantification of fractions of Ki-67 (h) or cleaved Caspase-3 (i) positive cells of the total number of tumour cells in mice treated as described in (e) and (f). n = 5 biological independent animals per BMI1High;CHD7Low vehicle-treated group, n = 4 biological independent animals per BMI1High;CHD7Low IP6 + Cisp-treated group, n = 2 biological independent animals per CTRL vehicle- and IP6 + Cisp-treated groups, one-way ANOVA. All graphs report mean ± SEM. P values: *P < 0.05, **P < 0.01, ***P < 0.001 or ****P < 0.0001. Scale bars = 100 µm. Source data are provided as a Source Data file.
Altogether, these data provide evidence of a synergic effect of IP6 and cisplatin in BMI1High;CHD7Low MB in vitro.
Combined IP6 and cisplatin treatment improves survival of BMI1High;CHD7Low MB xenografts
Leveraging the evidence that inositol administration through drinking water can rescue a brain-related lethal knockout phenotype51 and is therefore reaching the brain even when the blood–brain barrier (BBB) is intact, we set out to assess whether IP6 treatment could synergise with cisplatin also in a preclinical in vivo model of MB.
BMI1High;CHD7Low and control MB cells were injected into the cerebellum of newborn mice, which were treated with IP6 either alone or in combination with cisplatin at three weeks of age, with cisplatin being administered over a three-week period and IP6 until symptoms occurred (Fig. 5d). We observed significantly shorter survival in mice xenografted with BMI1High;CHD7Low MB as compared to control mice (Supplementary Fig. S5a), in keeping with our previous observation that patients with BMI1High;CHD7Low G4 MB have a poorer prognosis than other G4 MB patients10. Assessment of tumour size confirmed that these tumours were larger than control xenografts (Fig. 5g). Next, we examined the effect of the treatment on the survival of the mice xenografted with BMI1High;CHD7Low MB cells and found that both IP6 and cisplatin were significantly extending survival when compared to untreated mice, in particular the combination therapy led to longer survival as compared to either of the two single agents (IP6 or cisplatin alone) (Fig. 5e and Supplementary Fig. S5b, c). In contrast, xenografts derived from control cells benefited only from cisplatin treatment, with the combination treatment not being significantly different (Fig. 5f and Supplementary Fig. S5b, c). Analysis of the improvement in median survival, achieved in comparison to vehicle treatment, showed doubling of the survival of the BMI1High;CHD7Low xenografts compared to controls (without the signature) receiving the same treatment (Supplementary Fig. S5b–e). At a mechanistic level, impaired cell proliferation but no impact on apoptosis was found specifically in tumours originated from MB cells modelling the signature, as assessed by Ki-67 and cleaved Caspase-3 immunostaining (Fig. 5h, i), a finding that recapitulates the in vitro observation of impaired proliferation mediated by IP6 (Fig. 4a and Supplementary Fig. S4b).
In conclusion, these data confirm and extend our in vitro finding that IP6 enhances cisplatin cytotoxic activity only in MB cells modelling the BMI1High;CHD7Low signature and provide preclinical evidence that combinatorial treatment could be effective in patients with BMI1High;CHD7Low G4 MB.
Discussion
We have identified a novel epigenetic regulation of inositol metabolism in G4 MB with a BMI1High;CHD7Low molecular signature. We show overactivation of the mTOR pathway and induction of metabolic adaptation in G4 MB cells, but not in hNSC with the same signature, which can be counteracted by IP6 treatment. We provide proof of principle that IP6 synergises with cisplatin enhancing its cytotoxic activity in in vitro and in an in vivo pre-clinical model of BMI1High;CHD7Low MB where it significantly extended survival of the xenografted mice.
In a forward genetic screening approach in the mouse, we have previously described a molecular convergence of the chromatin remodellers BMI1 and CHD7, which also characterises a proportion of G4 MB with a significantly decreased overall survival10. We have shown overactivation of MAPK/ERK signalling as a downstream effector of this convergence mediating proliferation in MB cell lines and tumour growth in xenografts. Because increased proliferation and overactivation of the MAPK/ERK pathway were observed also in hNSC upon CHD7 silencing in a BMI1-overexpressing context10, it was conceivable that this was the mechanism underpinning neoplastic transformation of progenitors into a subset of G4 MB. Yet we show here that medulloblastoma did not develop upon modelling the signature in Math1+ progenitor cells in genetically engineered mice. Math1+ progenitor cells were chosen to test this hypothesis because originally transformed in the Sleeping Beauty-driven transposon screen10 and also because they have been proposed as the cell of origin of a proportion of G4 MB in single-cell transcriptomic studies comparing MB subgroup and progenitor cells during cerebellar development18,31. In particular, UBC progenitors which are derived from the Math1+ lineage are remarkably similar at the transcriptomic level to a proportion of G4 MB18,31. EOMES, a glutamatergic lineage-specific transcription factor previously described as a possible master regulator of G4 MB subgroup12, is expressed in UBC cells18,31. We show an increased number of EOMES+ UBC and reduced number of NeuN+ granule cells in Math1Cre;STOPFloxBmi1;Chd7f/+ mice suggesting that this signature, albeit not sufficient to elicit MB formation, favours a differentiation toward the UBC lineage rather than the granule cell lineage, potentially predisposing these cells to the acquisition of additional genetic/epigenetic lesions leading to malignant transformation.
BMI1 and CHD7 are chromatin remodelling factors known to modulate gene expression through epigenetic mechanisms52,53. Besides their traditional roles as members of the PRC1 complex54 and in the organisation of chromatin structure55, respectively, several reports have highlighted a direct role of PRC complexes in DNA methylation56,57,58,59 and CHD7 loss-of-function mutations have been associated with specific patterns of CpG DNA methylation modifications60. Therefore, to obtain a comprehensive understanding of the effects elicited by a BMI1High;CHD7Low signature in a neoplastic and non-neoplastic cell context, while also taking advantage of the extensive collection of publicly available omics datasets on bulk tumour tissue for comparative analysis, we studied the transcriptome and methylome of MB cells modelling the signature and integrated the results with those of the G4 bulk tumours. We show a striking convergence on phosphoinositol compounds metabolism when all datasets are integrated, indicating a mechanism shared between the cell model and the tumour bulk with a BMI1High;CHD7Low signature, which is consistent across in vitro and in vivo MB environmental conditions.
The identification of key mechanisms regulated by phosphorylation in BMI1High;CHD7Low G4 MB, together with the recent identification of a G4-specific phosphorylation profile61,62, prompted us to widen our analysis by analysing all phospho-regulated proteins in BMI1High;CHD7Low MB cells by mass spectrometry. We confirmed an enrichment in MAPK/ERK signalling, as expected10. However, we found that proteins regulated by phosphorylation in BMI1High;CHD7Low MB cells are significantly enriched for other RTK-related pathways as well, with activation of MET and FLT3 signalling being particularly highlighted. MET receptor has been described as a putative MB target63,64, although FLT3 has never been linked to MB before. MET and FLT3 are mTOR activators39,65,66. The role of mTOR in SHH canonical signalling is known67,68,69,70,71, hence inhibition of mTOR has been proposed as a therapeutic target for this subgroup, however, a potential role of mTOR in G4 MB is less well characterised. Transcriptome and methylome characterisation of G4γ subtype predicted activation of the PI3K/AKT/mTOR signalling pathway in this subgroup4, and integrative proteo-genomics analysis showed that RTK and downstream signalling constitute central oncogenic drivers in G4 MB62. Here we demonstrate that BMI1High;CHD7Low specifically enhances mTOR activation in MB and not in hNSC, a finding which is in agreement with MET and FLT3 not being deregulated in these cells, which are also not impacted by inhibition of FLT3. In the neoplastic context, our data support a specific impact on mTORC1 signalling. Indeed, while Torin, a broad inhibitor of both mTORC1 and mTORC2, partially affected the viability of MB cells without the signature, treatment with the mTORC1-specific inhibitor rapamycin only impacted on BMI1High;CHD7Low MB cells.
Both mTOR signalling and inositol contribute to the modulation of energy production42,43,44,45 which is crucial for the tumour to functionally adapt to a high proliferative state. We show a neoplastic-specific energetic adaptation in a BMI1High;CHD7Low context, with decreased mitochondrial function and concomitantly increased glycolytic activity, leading to increased ATP production in basal condition. This is a well-characterised phenomenon, known as the Warburg effect, which allows tumour cells to enhance aerobic glycolysis and use glycolytic intermediates to support macromolecule synthesis thereby sustaining proliferation in hypoxic condition20,72. Because both BMI1 and CHD7 have been previously linked to hypoxia, with BMI1 expression specifically found in the hypoxic region in glioblastoma73 and CHD7 being downregulated in response to low oxygen condition74, it is conceivable that a BMI1High;CHD7Low signature identifies a proportion of G4 MB with a specific advantage in hypoxic conditions due to the metabolic adaptation we have described. Notably, we observed enhanced expression of genes coding for enzymes involved in glucose metabolism and increased levels of valine and leucine by MRS in BMI1High;CHD7Low G4 MB tissues confirming our conclusions from the in vitro data and raising the possibility that imaging biomarkers could be developed to identify patients with the signature. Interestingly, we show that the same signature in hNSC does not elicit the same effect, despite the increased proliferative activity acquired by the cells, raising the possibility that additional events leading to the acquisition of metabolic adaptation are essential for neoplastic transformation.
IP6 antitumour activity has been documented in different types of tumour cell lines and is mainly mediated by cell growth inhibition27,75; however, its role in MB had never been investigated. We show that IP6 treatment counteracted both mTOR activation and metabolic adaptation in BMI1High;CHD7Low MB cells and inhibited their proliferation. Importantly, IP6 sensitivity in BMI1High;CHD7Low MB cells is mediated by a novel BMI-dependent epigenetic regulation at promoters of genes involved in PLC activation, and thus lipid inositol signalling, where co-occupancy by BMI1 and H3K27me3 is demonstrated specifically in BMI1High;CHD7Low MB cells. These data are in agreement with a previous observation in embryonic stem cells where genes related to inositol-phosphate metabolism were identified as PRC targets76. Interestingly, we also found that BMI1 co-localises at bivalent domains, marked H3K27me3 and H3K4me3, in MB cells and in particular onto the promoter of the PPIP5K2 gene, a kinase responsible for phosphorylation of IP6 to higher inositol products, which is specifically downregulated in BMI1High;CHD7Low MB cells upon inositol treatment.
Our data raise the possibility that a BMI1-mediated regulation at the PPIP5K2 promoter allows BMI1High;CHD7Low cells to reduce its expression upon dysregulation of inositol metabolism, therefore preventing IP6 conversion and enhancing its cytotoxic effect. In keeping with this interpretation, silencing of PPIP5K2 led to decreased cell proliferation in colon cancer cells77. Future studies will be required to elucidate the role of PPIP5K2 in MB metabolism and growth and to investigate its potential role as a predictive biomarker, once its methylation status in BMI1High;CHD7Low G4 MB patients will be tested; something not possible in publicly available datasets as the gene is not present in the DNA methylation arrays used for these studies. Moreover, we show that MET and FLT3 promoters are differentially bound by BMI1 upon CHD7 silencing, in agreement with loss of BMI1-dependent repression of these RTKs in BMI1High;CHD7Low MB cells that in turn become more sensitive to their inhibition and show enhanced mTOR pathway activation.
Cancer treatment frequently benefits from a combination therapy, which relies on targeting multiple pathways to obtain a cooperative and superior effect while reducing drug resistance78. Adjuvant inositol treatment has been shown to attenuate the side effects of chemotherapy in breast and colon cancer79. We reasoned that combination therapy of cisplatin and IP6 could be effective in BMI1High;CHD7Low G4 MB because of the overactivation of the mTOR pathway upon cisplatin treatment80,81, which can be counteracted by IP6 and the enhancement of cisplatin-mediated antitumoral effect by inhibition of glycolysis82,83, which can also be induced by IP6. Our results show that IP6 treatment acts synergistically with cisplatin in BMI1High;CHD7Low MB cells both in vitro and in an in vivo xenograft model and suggest that IP6 administration could be implemented as adjuvant therapy in G4 MB patients that are currently treated with cisplatin5. Moreover, we did not observe toxicity of IP6 on hNSC or side effects upon administration to mice, in keeping with previous reports where normal bone marrow progenitor cells84 and peripheral blood mononuclear cells and T-cell colony-forming cells are unaffected by inositol compound treatment27.
In conclusion, deconvolution of the molecular convergence of the chromatin remodellers BMI1 and CHD7 in MB, uncovered a novel epigenetic regulation of phosphoinositol metabolism, which mediates a metabolic energetic adaptation in BMI1High;CHD7Low subgroup of G4 MB and predicts response to inositol pathway modulation in combination with chemotherapeutics currently used to treat patients with MB. We, therefore, identify a potentially actionable vulnerability of this particular group of patients.
Methods
Cell culture conditions
ICb1299 patient-derived MB lines were obtained from Dr Xiao-Nan Li, Baylor College of Medicine, Texas Children Cancer Centre, USA85. ICb1299 was cultured in DMEM (high glucose, GlutaMAX™, ThermoFisher) supplemented with 10% foetal bovine serum (FBS, Gibco) and 1% penicillin–streptomycin (Gibco). CHLA-01-Med MB cells were purchased from ATCC (CRL3021) and cultured as described previously86. Briefly, cells were grown in DMEM-F12 (Gibco) supplemented with B27 (Gibco), 20 ng/mL human epidermal growth factor (EGF, Peprotech) and 10 ng/mL human basic fibroblast growth factor (FGF, Peprotech). MB subtype identity for the two cell lines has been previously shown10,85.
Human foetal NSC lines were obtained from the Cancer Research UK-funded Glioma Cellular Genetics Resource (www.gcgr.org.uk) and cultured as previously described87. Informed consent was obtained from all donors of tissue for NSC derivation. Ethical approval was obtained from South East Scotland REC (Reference 08/S1101/1 and Amendment 08/S1101/1/AM06/1) and management approval was obtained from Lothian NHS Board. Cells were maintained at 37 °C and were sub-cultured every 3 days once they reached confluence.
Cell viability, growth and apoptosis assays
For growth curve and cell viability assays, MB cells or hNSCs were seeded in 24-well plates at the same density. Cells were treated for 72 h with mTOR (rapamycin, Sigma; Torin1, Calbiochem), FLT3 (TCS359, SelleckChem) or MET (PHA665752, Tocris) inhibitors or with inositol hexakisphosphate (IP6, Sigma) at the indicated concentrations. At specific time points or after appropriate treatment, cells were harvested, and the number of viable cells was counted with a haemocytometer and Trypan Blue staining or with CyQUANT Direct Red Cell Proliferation Assay Kit (ThermoFisher Scientific). Syenrgy/antagonist effect of the combined treatment was identified by Loewe Model with Combenefit software88. Apoptosis was assessed with Caspase-3 Colorimetric Assay kit (ab39401, Abcam) following the manufacturer’s protocol. Briefly, cells treated with increasing concentrations of IP6 were lysed, and 50 µg of protein lysate was incubated with DEVD-AFC substrate and reaction buffer for 2 h at 37 °C.
Production of shRNA lentiviral vector and gene silencing
GIPZ lentiviral shRNA vectors containing a hairpin sequence targeting BMI1 or CHD7 and the coding sequences for GFP and puromycin-resistance gene were purchased from Dharmacon, UK. Packaging, virus production and determination of titre were carried out as previously reported10. Cells were infected overnight at a multiplicity of infection (MOI) of 1. After 96 h from the infection, puromycin selection at a concentration of 2.5 µg/ml was applied to enrich the transduced population. The efficacy of gene silencing was assessed by qRT-PCR or western blot analysis.
RNA extraction and qRT-PCR analysis
The total RNA was isolated from cell pellets with RNeasy Micro purification kit (Qiagen) and digested with DNaseI (Applied Biosystems). The cDNA synthesis was carried out with SuperScript III Reverse Transcriptase Kit (Invitrogen) following the manufacturer’s protocol. Analysis of gene expression was performed with the Applied Biosystems 7500 Real-Time PCR System using TaqMan gene expression MasterMix (Applied Biosystems) and SYBR Green PCR MasterMix (Applied Biosystems) according to standard protocols. Technical triplicates for each sample were analysed. The Ct values of all the genes analysed were normalised to the Ct of GAPDH for TaqMan probes or average Ct of ACTB and ATP5F1B for SYBR Green PCR and fold changes were calculated. Target genes and TaqMan probes used are the following: EOMES, Hs00172872_m1; MT-ND1, Hs02596873_s1; GAPDH, Hs02758991_g1. Primers used in SYBR Green qPCR are listed in Supplementary Table 1.
Western blot analysis
MB cells or hNSCs were lysed for 30 min on ice using RIPA lysis buffer supplemented with 2 mM PMSF, 1 mM sodium orthovanadate and protease inhibitor cocktail (PIC) (RIPA Lysis Buffer System, Santa Cruz) followed by three pulses of sonication to obtain total protein lysate. Nuclear and cytoplasmic fractions of cells were obtained with two different lysis buffers as described previously89. Briefly, cells were harvested in Buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.15% Nonidet P40 (NP40) and 0.1 mM EGTA) supplemented with 1 mM DTT and PIC. Cells were homogenised through a 26-G needle, nuclei were isolated by centrifugation and the supernatant (cytoplasmic fraction) was collected. Nuclei were washed quickly with ice-cold PBS, suspended in Buffer B (20 mM HEPES, pH 7.9, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA and 0,5% NP40) supplemented with 1 mM DTT and PIC and lysed by sonication. Protein concentration was determined using BCA Protein Assay Kit (Pierce). Equal amounts of protein were separated by SDS-PAGE and transferred onto a nitrocellulose membrane (Amersham). After transfer, the membrane was blocked for an hour at room temperature in 5% skimmed milk in TBST buffer (25 mM Tris-HCl, 137 mM NaCl and 0.1% Tween 20, pH 7.5) and probed with different antibodies. Incubation with primary antibody was performed overnight at 4 °C followed by appropriate secondary HRP-conjugated antibodies (anti-rabbit IgG or anti-mouse IgG, 1:5000, Amersham) for one hour at room temperature. Enhanced chemiluminescence (ECL Plus, Amersham) was used for the detection of the bands. The following primary antibodies were used: mouse monoclonal anti-BMI1 (1:1000, clone AF27, Active Motif), anti-GAPDH (1:1000, G8795, Sigma) and anti-Vinculin (1:5000, V4505, Sigma), rabbit monoclonal anti-phospho-RPS6 (Ser240/244, 1:1000, D68F8, Cell Signaling), anti-4EBP1 (1:1000, 53H11, Cell Signaling), anti-phospho-4EBP1 (Thr37/46, 1:1000, 236B4, Cell Signaling), rabbit polyclonal anti-CHD7 (1:500, ab117522, Abcam), anti-RPS6 (1:1000, 5G10, Cell Signaling), anti-PPIP5K2 (1:500, 16836, Novus Biological) and goat polyclonal anti-Lamin B (1:5000, sc-6216, Santa Cruz).
Mytotracker and immunofluorescence analysis
MB or hNSC cells, incubated for 1 h at 37 °C with 0.02 nM MitoTracker Red CMXRos (ThermoFisher Scientific), were fixed using 4% PFA, permeabilised with 0.2% Triton® X-100 and mounted for immunofluorescence analysis. Relative red fluorescent signal from mitotracker was quantified with ImageJ.
OCR and ECAR analysis
Metabolic extracellular flux was measured using Seahorse Extracellular Flux Analyzer XF-24 (Agilent Technology) as previously described90. Briefly, 1.7 × 105 MB or hNSC cells were seeded on XF-24 cell culture microplates, coated with 3.4 mg/mL BD Cell-TakTM tissue adhesive solution (BD Bioscience 354240), on the same day of the analysis. For OCR and ECAR analysis after IP6 treatment, cells were incubated with IP6 for 24 h before seeding on coated wells. Microplates containing the cell suspension were centrifuged at 180×g for 2 min and then incubated at 37 °C with unbuffered DMEM (Sigma) in a non-CO2 incubator for at least 2 h. OCR was determined following sequential treatment with the ATPase inhibitor oligomycin (5 µM), the uncoupling agent FCCP (4 µM) and the electron-transport-chain inhibitors rotenone (10 µM) and antimycin A (10 µM). ECAR was measured in response to glucose (10 mM), oligomycin (5 µM) and 2DG (50 mM) treatment.
Total ATP and ADP/ATP ratio measurements
1 × 105 MB or hNSC cells were cultured for 24 h and then harvested and washed with ice-cold PBS to remove extracellular ATP. The cell pellet was resuspended in ice-cold water and sonicated for 5 min with cycles of 30 s on and 30 s off. 25 µl of cell lysate were incubated with 25 µl of CellTiter-Glo Luminescent Cell Viability Assay solution (Promega) for 10 min. ADP/ATP ratio was measured with ADP/ATP Ratio Assay Kit (Sigma Aldrich) following the manufacturer’s protocol. The luminescence intensity was measured using a Synergy HT Multi-Mode Microplate Reader, normalised on the total amount of proteins and expressed as RLU (relative luminescence unit)/µg of proteins.
Mass spectrometry sample preparation and LC–MS/MS analysis
Protein extraction and trypsin digestion were performed as previously described91,92. Peptide solutions were desalted using 10 mg of OASIS-HLB cartridges (Waters, UK) and peptides were eluted with glycolic acid buffer (1 M glycolic acid, 50% ACN, 5% TFA). To enrich phosphopeptides, sample volumes were normalised using glycolic acid buffer (1 M glycolic acid, 80% ACN, 5% TFA) and TiO2 beads ((50% slurry in 1% TFA), Hichrom).
For LC–MS/MS analysis, phosphopeptides were reconstituted in 13 µL of reconstitution (97% H2O, 3% ACN and 0.1% TFA, 50 fmol/µl enolase peptide digest) and sonicated for 2 min at RT. Phosphopeptides were analysed by nanoflow ultimate 3000 RSL nano instrument that was coupled online to a Q-Exactive plus mass spectrometer (ThermoFisher Scientific). Gradient elution was from 3 to 35% buffer B in 120 min at a flow rate of 250 nL/min with buffer A being used to balance the mobile phase (buffer A was 0.1% formic acid in water and B was 0.1% formic acid in ACN). The mass spectrometer was controlled by Xcalibur software (version 4.0) and operated in the positive mode. The spray voltage was 1.95 kV and the capillary temperature was set to 255 °C. The Q-Exactive plus was operated in data-dependent mode with one-survey MS scan followed by 15 MS/MS scans. The full scans were acquired in the mass analyser at 375–1500 m/z with a resolution of 70,000, and the MS/MS scans were obtained with a resolution of 17,500.
Peptide identification and quantification
For peptide identification, MS raw files were converted into Mascot Generic Format using Mascot Distiller (version 2.7.1) and searched against the Swiss-Prot database (release September 2019) (https://www.uniprot.org/statistics/Swiss-Prot) restricted to human entries using the Mascot search daemon (Ref 2, version 2.6.0) with a FDR of ~1% and restricted to the human entries. Allowed mass windows were 10 ppm and 25 mmu for parent and fragment mass to charge values, respectively. Variable modifications included in searches were oxidation of methionine, pyro-glu (N-term) and phosphorylation of serine, threonine and tyrosine. Peptides with an expectation value <0.05 were considered for further analysis.
The mascot result (DAT) files were extracted into excel files. For peptide quantification, Pescal Software [3] was used to construct extracted ion chromatograms (XIC) for all the identified peptides across all conditions and calculating the peak heights. These peptide peak heights were then normalised to the sum of the intensities for each individual sample and the average fold change between conditions could be determined. Statistical significance between conditions was considered significant when the Student’s T tests produced P < 0.05 following Benjamini–Hochberg (BH) multiple testing correction. Kinase substrate enrichment analysis (KSEA) was performed as described before93. Briefly, peptides differentially phosphorylated between a set of samples (at non-adjusted P < 0.05) were grouped into substrate sets known to be phosphorylated by a specific kinase as annotated in the PhosphoSite (https://www.phosphosite.org/), Phospho.ELM (http://phospho.elm.eu.org/) and PhosphoPOINT (http://kinase.bioinformatics.tw/) databases94,95,96,97. To infer the enrichment of substrate groups across sets of samples, the hypergeometric test was used, followed by BH multiple testing correction.
DNA methylation analysis
Total genomic DNA was extracted with RNA/DNA/Protein Purification Plus kit (Norgen), and DNA methylation data were assayed on the Illumina Human Methylation EPIC microarray after bisulfite conversion. Raw data were preprocessed using the Bioconductor ChAMP package in R to remove failed detections and probes with design flaws98. Normalisation was performed using the SWAN algorithm99. Probes were mapped to genes using the methylation array annotation. One probe may map to multiple genes. These operations were carried out on both cell lines simultaneously, after which they were analysed separately. Two preprocessing steps were then applied to increase our ability to detect differential methylation. First, any probes whose M values had a range less than 1 M value unit across all experimental groups were removed, then any probes whose M values varied by greater than 5 M value units within a single experimental group were removed. Differentially methylated probes were identified using the limma package in R100. Probes with a false discovery rate <0.05 were declared differentially methylated.
RNA sequencing (RNA-Seq) analysis
The total RNA was extracted with RNA/DNA/Protein Purification Plus kit (Norgen), polyA mRNA was selected and library-prepared using the NEBNext Ultra II Directional RNA Library Prep kit. Libraries were size-selected, multiplexed and 75-bp paired-end sequences were obtained with HiSeq4000 (Illumina). Quality control was performed with FastQC and adapter sequences were removed using Trimgalore v0.6.5 (www.bioinformatics.babraham.ac.uk/projects/). Reads were then aligned to the Ensembl GRCh38 human reference genome using STAR v2.6.1101, with gene count quantification mode, retaining only uniquely mapped reads. Lowly expressed genes (CPM < 1) were filtered out via a custom-made script in R, and further biotype filtering was performed in R with the Bioconductor packages NOISeq102 and biomaRt103, removing residual highly expressed mitochondrial and ribosomal RNA. Trimmed mean of M-values (TMM) normalisation was applied to the dataset prior to differential expression (DE) analysis, performed using the Bioconductor package edgeR104. Specifically, a negative binomial generalised log-linear model (glmfit) was fitted to the read counts and the likelihood ratio test (glmLRT) was conducted for each comparison of interest. The Benjamini–Hochberg FDR cut-off was set at 0.05. The versions of all relevant Bioconductor packages were compatible with R v3.5.3.
Chromatin immunoprecipitation (ChIP) assays
BMI1, H3K4me3 and H3K27me3 ChIPs were performed using ChIP-IT High Sensitivity kit (Active Motif) following the manufacturer’s instructions. Briefly, 1 × 106 cells were fixed with the formaldehyde-based fixing solution for 15 min at room temperature and lysed with provided lysis solution supplemented with protease inhibitors. Next, nuclei pellets were lysed and chromatin sonicated with Bioruptor Plus sonication device (Diagenode) to obtain DNA fragments within the recommended 200–1200-bp range. In total, 25 µg or 10 µg of sheared chromatin was then incubated with 4 µg of antibody against BMI1 (BMI1, clone AF27, Active Motif), H3K27me3 (Diagenode) or H3K4me3 (Diagenode) overnight at 4 °C with rotation. Following incubation with Protein G agarose beads, bound chromatin was washed, eluted and purified following the manufacturer’s protocols. Validation by qPCR-ChIP on target genes was done before proceeding to sequencing. ChIPed DNA was end-repaired, A-tailed and adapter-ligated before size selection and amplification. The obtained libraries were QC’ed and multiplexed before 75-bp paired-end sequencing on HiSeq4000 (Illumina).
ChIP-Seq analysis
The quality of ChIP-Seq samples was first assessed via FastQC and TrimGalore, removing low-quality and adapter sequences. The average Phred score of the surviving reads across all samples was 30 and the average sequencing depth was 36.3 M (min = 22.1 M, max = 56.7 M). The alignment to the Ensembl GRCh38 human reference genome was performed via Bowtie v2.3.4105 with default parameters, in concomitance with the usage of samtools106 for the post processing and sorting of the Binary Alignment Map (bam) files. Exploratory tools such as deeptools107, plotCorrelation, plotPCA and plotFingerprint on Python v2.7.15 were used to further assess sample characteristics and to address the potential presence of outliers. After performing post-alignment quality checks, peaks were called via the MACS2 algorithm (subroutine callpeak)108 against the corresponding input background. The shifting model was disabled to make different datasets comparable and the “--broad” option was enabled for the analysis of the histone mark H3K27me3. A minimum fold enrichment of 2 was selected with an FDR of 0.05, in both narrow peak and broad peak (--broad-cut-off) statistical analyses. The Bioconductor packages in R GenomicRanges109 and ChIPSeeker110 were used to find regions of consensus peaks between the two cell lines, for each antibody/condition, and to annotate them based on the location with respect to the nearest transcription start site (TSS): promoters (within 3 Kb from the TSS), exons, introns, 5′ UTR, 3′ UTR and distal intergenic. The latter was merged with so-called downstream regions. The versions of all relevant Bioconductor packages were compatible with R v3.5.3.
ChIP-Seq heatmap generation
The deeptools algorithms bamCompare, bigWigCompare and plotHeatmap were used to produce relevant bigwig files and heatmaps, in order to assess region-wide and genome-wide coverage. Specifically, coverage tracks were first obtained by normalising each sample alignment file against the corresponding input (--operation log2ratio) and then pooled at both replicate and cell line levels (--operation mean), to obtain a representative sample for each antibody/condition. The regions chosen to be visualised in the heatmaps were those of consensus peaks (details in the figure legends), centred on their nearest TSS.
Pathway and gene set analysis
Ingenuity pathway analysis (IPA, Qiagen), Reactome (https://reactome.org/)111 and Panther Gene Ontology tool (http://geneontology.org/) were used to assess biological pathways and gene ontology terms that showed significant enrichment in the various gene sets. Functional analysis of significant DE and DMP gene lists obtained for MB cells or tumour samples was carried out with IPA software with canonical pathways tool following the manufacturer’s standard protocol. Pathways that were significant in both RNA-Seq and DNA methylation analysis and conserved between cells and tumour samples were visualised by Venn diagrams. Reactome Pathway Database was used to interrogate datasets of differentially phosphorylated proteins, while Panther Gene Ontology was used to analyse GO terms significantly enriched in ChIP-Seq analysis. The enrichment for each term was deemed statistically significant if the adjusted P value (FDR) was lower than 0.05.
Cytoscape v. 3.7.2112 was used to visualise relevant biological networks of enriched pathways, together with EnrichmentMap and AutoAnnotate applications. Several layout parameters were tuned to achieve the current Cytoscape visualisation.
Genetically engineered mouse model
Math1Cre;STOPFloxBmi1;Chd7f/+, Math1Cre;STOPFloxBmi1 and STOPFloxBmi and Chd7f/+ control mice were generated by crossing Math1Cre mice or Math1Cre;Chd7f/f mice with STOPFloxBmi1 mice. Mouse lines were as described previously113,114,115. In vivo BrdU labelling and analyses were performed as described previously114. For immunostainings, brains were dissected, fixed in 4% PFA overnight and processed for immunostaining as previously described10, using rabbit anti-EOMES (Sigma Aldrich).
Animal experiments
All procedures were performed in accordance with licences held under the UK Animals (Scientific Procedures) Act 1986 and later modifications and conforming to all relevant guidelines and regulations. Mice were kept at ~18–23 °C with 40–60% humidity in a 12-h light/12-h dark cycle. Required sample sizes were calculated by an a priori power analysis and mice were assigned randomly to the different groups of treatment. 1 × 105 MB cells were injected into the right cerebellar hemisphere of P5 mice. After 2 weeks from the injection, mice were treated with cisplatin and/or IP6. Cisplatin (Sigma) was administered by intraperitoneal (i.p.) injection at 3.5 mg/kg in saline solution. 2% IP6 (Sigma) was administered in sterile drinking water, pH-adjusted to 7.4 for oral administration and replaced every 48 h. Control mice were i.p.-injected with saline solution, as the vehicle.
Data analysis and statistical methods
The number of experiments (n) used for all the statistical tests is specified in each figure legend. All quantitative data are presented as mean ± standard error of the mean (SEM) of at least three experiments, unless otherwise specified. The statistical significance was determined by two-tailed unpaired Student’s t test, one-way ANOVA followed by Tukey’s multiple-comparison test or two-way ANOVA followed by Dunnet’s multiple-comparison test. Survival of xenografted mice was estimated with Kaplan–Meier survival analysis, and significance was determined with Log-rank (Mantel–Cox) test. Statistical analysis was performed using GraphPad software and statistical significance is represented as *P < 0.05, **P < 0.01, ***P < 0.001 or ****P < 0.0001.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The authors declare that all the data supporting the findings of this study are available within the article and its Supplementary Information files. The datasets generated in this study and processed data are available in the NCBI Gene Expression Omnibus database (GSE156077) or are available from the corresponding author upon reasonable request. Publicly available datasets used in the study: GSE85217 (Expression data from primary medulloblastoma samples), Reactome (https://reactome.org/), Panther Gene Ontology tool (http://geneontology.org/), Swiss-Prot (https://www.uniprot.org/statistics/Swiss-Prot), PhosphoSite (https://www.phosphosite.org/), Phospho.ELM (http://phospho.elm.eu.org/) and PhosphoPOINT (http://kinase.bioinformatics.tw/). Source data are provided with this paper.
Code availability
The custom-made code used in this study is available in the GitHub repository (https://github.com/nickpom88/rnaseq/blob/main/filter_genes_np2.R).
References
Northcott, P. A. et al. Medulloblastoma comprises four distinct molecular variants. J. Clin. Oncol. 29, 1408–1414 (2011).
Taylor, M. D. et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 123, 465–472 (2012).
Schwalbe, E. C. et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol. 18, 958–971 (2017).
Cavalli, F. M. G. et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell 31, 737–754 (2017). e736.
Mueller, S. & Chang, S. Pediatric brain tumors: current treatment strategies and future therapeutic approaches. Neurotherapeutics 6, 570–586 (2009).
Jones, B., Wilson, P., Nagano, A., Fenwick, J. & McKenna, G. Dilemmas concerning dose distribution and the influence of relative biological effect in proton beam therapy of medulloblastoma. Br. J. Radiol. 85, e912–e918 (2012).
Pugh, T. J. et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature 488, 106–110 (2012).
Robinson, G. et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 488, 43–48 (2012).
Dubuc, A. M. et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol. 125, 373–384 (2013).
Badodi, S. et al. Convergence of BMI1 and CHD7 on ERK signaling in medulloblastoma. Cell Rep. 21, 2772–2784 (2017).
Behesti, H., Bhagat, H., Dubuc, A. M., Taylor, M. D. & Marino, S. Bmi1 overexpression in the cerebellar granule cell lineage of mice affects cell proliferation and survival without initiating medulloblastoma formation. Dis. Model Mech. 6, 49–63 (2013).
Lin, C. Y. et al. Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature 530, 57–62 (2016).
Gargiulo, G. et al. In vivo RNAi screen for BMI1 targets identifies TGF-beta/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell 23, 660–676 (2013).
Leung, C. et al. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 428, 337–341 (2004).
Bruggeman, S. W. et al. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell 12, 328–341 (2007).
Glinsky, G. V., Berezovska, O. & Glinskii, A. B. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J. Clin. Investig. 115, 1503–1521 (2005).
Merve, A. et al. Polycomb group gene BMI1 controls invasion of medulloblastoma cells and inhibits BMP-regulated cell adhesion. Acta Neuropathol. Commun. 2, 10 (2014).
Vladoiu, M. C. et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572, 67–73 (2019).
Miranda-Goncalves, V., Lameirinhas, A., Henrique, R. & Jeronimo, C. Metabolism and epigenetic interplay in cancer: regulation and putative therapeutic targets. Front. Genet. 9, 427 (2018).
Venneti, S. & Thompson, C. B. Metabolic reprogramming in brain tumors. Annu Rev. Pathol. 12, 515–545 (2017).
Goel, A., Mathupala, S. P. & Pedersen, P. L. Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. J. Biol. Chem. 278, 15333–15340 (2003).
Pan, D., Mao, C. & Wang, Y. X. Suppression of gluconeogenic gene expression by LSD1-mediated histone demethylation. PLoS ONE 8, e66294 (2013).
Wolf, A., Agnihotri, S., Munoz, D. & Guha, A. Developmental profile and regulation of the glycolytic enzyme hexokinase 2 in normal brain and glioblastoma multiforme. Neurobiol. Dis. 44, 84–91 (2011).
Moreno-Sanchez, R., Rodriguez-Enriquez, S., Saavedra, E., Marin-Hernandez, A. & Gallardo-Perez, J. C. The bioenergetics of cancer: is glycolysis the main ATP supplier in all tumor cells? Biofactors 35, 209–225 (2009).
Chakraborty, A. et al. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell 143, 897–910 (2010).
Prasad, A. et al. Inositol hexakisphosphate kinase 1 regulates neutrophil function in innate immunity by inhibiting phosphatidylinositol-(3,4,5)-trisphosphate signaling. Nat. Immunol. 12, 752–760 (2011).
Bizzarri, M., Dinicola, S., Bevilacqua, A. & Cucina, A. Broad spectrum anticancer activity of myo-inositol and inositol hexakisphosphate. Int. J. Endocrinol. 2016, 5616807 (2016).
Ferry, S., Matsuda, M., Yoshida, H. & Hirata, M. Inositol hexakisphosphate blocks tumor cell growth by activating apoptotic machinery as well as by inhibiting the Akt/NFkappaB-mediated cell survival pathway. Carcinogenesis 23, 2031–2041 (2002).
Karmakar, S., Banik, N. L. & Ray, S. K. Molecular mechanism of inositol hexaphosphate-mediated apoptosis in human malignant glioblastoma T98G cells. Neurochem. Res. 32, 2094–2102 (2007).
Vucenik, I. et al. Anti-angiogenic activity of inositol hexaphosphate (IP6). Carcinogenesis 25, 2115–2123 (2004).
Hovestadt, V. et al. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature 572, 74–79 (2019).
Weyer, A. & Schilling, K. Developmental and cell type-specific expression of the neuronal marker NeuN in the murine cerebellum. J. Neurosci. Res. 73, 400–409 (2003).
Rusert, J. M. et al. Functional precision medicine identifies new therapeutic candidates for medulloblastoma. Cancer Res. 80, 5393–5407 (2020).
Xu, J. et al. Disseminated medulloblastoma in a child with germline BRCA2 6174delT mutation and without fanconi anemia. Front. Oncol. 5, 191 (2015).
Zhao, X. et al. Global gene expression profiling confirms the molecular fidelity of primary tumor-based orthotopic xenograft mouse models of medulloblastoma. Neuro Oncol. 14, 574–583 (2012).
Kumar, R. et al. Clinical outcomes and patient-matched molecular composition of relapsed medulloblastoma. J. Clin. Oncol. 39, 807–821 (2021).
Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010).
Hubbard, S. R. & Miller, W. T. Receptor tyrosine kinases: mechanisms of activation and signaling. Curr. Opin. Cell Biol. 19, 117–123 (2007).
Chen, W. et al. mTOR signaling is activated by FLT3 kinase and promotes survival of FLT3-mutated acute myeloid leukemia cells. Mol. Cancer 9, 292 (2010).
Demkova, L. & Kucerova, L. Role of the HGF/c-MET tyrosine kinase inhibitors in metastasic melanoma. Mol. Cancer 17, 26 (2018).
Lam, B. Q., Dai, L. & Qin, Z. The role of HGF/c-MET signaling pathway in lymphoma. J. Hematol. Oncol. 9, 135 (2016).
Bridges, D. & Saltiel, A. R. Phosphoinositides: key modulators of energy metabolism. Biochim Biophys. Acta 1851, 857–866 (2015).
Szijgyarto, Z., Garedew, A., Azevedo, C. & Saiardi, A. Influence of inositol pyrophosphates on cellular energy dynamics. Science 334, 802–805 (2011).
de la Cruz Lopez, K. G., Toledo Guzman, M. E., Sanchez, E. O. & Garcia Carranca, A. mTORC1 as a regulator of mitochondrial functions and a therapeutic target in cancer. Front. Oncol. 9, 1373 (2019).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017).
Bennett, C. D. et al. Ex vivo metabolite profiling of paediatric central nervous system tumours reveals prognostic markers. Sci. Rep. 9, 10473 (2019).
Gresset, A., Sondek, J. & Harden, T. K. The phospholipase C isozymes and their regulation. Subcell. Biochem. 58, 61–94 (2012).
Owusu Obeng, E. et al. Phosphoinositide-dependent signaling in cancer: a focus on phospholipase C isozymes. Int. J. Mol. Sci. 21, 2581 (2020).
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Fridy, P. C., Otto, J. C., Dollins, D. E. & York, J. D. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J. Biol. Chem. 282, 30754–30762 (2007).
Buccafusca, R. et al. Characterization of the null murine sodium/myo-inositol cotransporter 1 (Smit1 or Slc5a3) phenotype: myo-inositol rescue is independent of expression of its cognate mitochondrial ribosomal protein subunit 6 (Mrps6) gene and of phosphatidylinositol levels in neonatal brain. Mol. Genet. Metab. 95, 81–95 (2008).
Murawska, M. & Brehm, A. CHD chromatin remodelers and the transcription cycle. Transcription 2, 244–253 (2011).
Aranda, S., Mas, G. & Di Croce, L. Regulation of gene transcription by Polycomb proteins. Sci. Adv. 1, e1500737 (2015).
Gil, J. & O’Loghlen, A. PRC1 complex diversity: where is it taking us? Trends Cell Biol. 24, 632–641 (2014).
Basson, M. A. & van Ravenswaaij-Arts, C. Functional insights into chromatin remodelling from studies on CHARGE syndrome. Trends Genet. 31, 600–611 (2015).
Gao, F. et al. DNA methylation in the malignant transformation of meningiomas. PLoS ONE 8, e54114 (2013).
Laugesen, A., Hojfeldt, J. W. & Helin, K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. Mol. Cell 74, 8–18 (2019).
Li, Y. et al. Genome-wide analyses reveal a role of Polycomb in promoting hypomethylation of DNA methylation valleys. Genome Biol. 19, 18 (2018).
Vire, E. et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439, 871–874 (2006).
Butcher, D. T. et al. CHARGE and Kabuki Syndromes: gene-specific DNA methylation signatures identify epigenetic mechanisms linking these clinically overlapping conditions. Am. J. Hum. Genet. 100, 773–788 (2017).
Archer, T. C. et al. Proteomics, post-translational modifications, and integrative analyses reveal molecular heterogeneity within medulloblastoma subgroups. Cancer Cell 34, 396–410 (2018). e398.
Forget, A. et al. Aberrant ERBB4-SRC signaling as a hallmark of group 4 medulloblastoma revealed by integrative phosphoproteomic profiling. Cancer Cell 34, 379–395 (2018). e377.
Kongkham, P. N., Onvani, S., Smith, C. A. & Rutka, J. T. Inhibition of the MET receptor tyrosine kinase as a novel therapeutic strategy in medulloblastoma. Transl. Oncol. 3, 336–343 (2010).
Guessous, F. et al. An orally bioavailable c-Met kinase inhibitor potently inhibits brain tumor malignancy and growth. Anticancer Agents Med. Chem. 10, 28–35 (2010).
Hervieu, A. & Kermorgant, S. The role of PI3K in Met driven cancer: a recap. Front. Mol. Biosci. 5, 86 (2018).
Lindblad, O. et al. Aberrant activation of the PI3K/mTOR pathway promotes resistance to sorafenib in AML. Oncogene 35, 5119–5131 (2016).
Wu, C. C. et al. mTORC1-mediated inhibition of 4EBP1 is essential for hedgehog signaling-driven translation and medulloblastoma. Dev. Cell 43, 673–688 (2017). e675.
Pocza, T. et al. mTOR pathway as a potential target in a subset of human medulloblastoma. Pathol. Oncol. Res. 20, 893–900 (2014).
Rao, G. et al. Sonic hedgehog and insulin-like growth factor signaling synergize to induce medulloblastoma formation from nestin-expressing neural progenitors in mice. Oncogene 23, 6156–6162 (2004).
Eckerdt, F. et al. Pharmacological mTOR targeting enhances the antineoplastic effects of selective PI3Kalpha inhibition in medulloblastoma. Sci. Rep. 9, 12822 (2019).
Cancer, M. et al. Humanized stem cell models of pediatric medulloblastoma reveal an Oct4/mTOR axis that promotes malignancy. Cell Stem Cell 25, 855–870 (2019). e811.
Zhang, Y. & Yang, J. M. Altered energy metabolism in cancer: a unique opportunity for therapeutic intervention. Cancer Biol. Ther. 14, 81–89 (2013).
Jin, X. et al. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat. Med. 23, 1352–1361 (2017).
Boyd, N. H. et al. Chromodomain helicase DNA-binding protein 7 is suppressed in the perinecrotic/ischemic microenvironment and is a novel regulator of glioblastoma angiogenesis. Stem Cells 37, 453–462 (2019).
Vucenik, I. & Shamsuddin, A. M. Cancer inhibition by inositol hexaphosphate (IP6) and inositol: from laboratory to clinic. J. Nutr. 133, 3778S–3784S (2003).
Brookes, E. et al. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell 10, 157–170 (2012).
Gu, C. et al. KO of 5-InsP7 kinase activity transforms the HCT116 colon cancer cell line into a hypermetabolic, growth-inhibited phenotype. Proc. Natl Acad. Sci. USA 114, 11968–11973 (2017).
Bayat Mokhtari, R. et al. Combination therapy in combating cancer. Oncotarget 8, 38022–38043 (2017).
Bacic, I., Druzijanic, N., Karlo, R., Skific, I. & Jagic, S. Efficacy of IP6 + inositol in the treatment of breast cancer patients receiving chemotherapy: prospective, randomized, pilot clinical study. J. Exp. Clin. Cancer Res. 29, 12 (2010).
Liang, S. Q. et al. mTOR mediates a mechanism of resistance to chemotherapy and defines a rational combination strategy to treat KRAS-mutant lung cancer. Oncogene 38, 622–636 (2019).
Leisching, G. R., Loos, B., Botha, M. H. & Engelbrecht, A. M. The role of mTOR during cisplatin treatment in an in vitro and ex vivo model of cervical cancer. Toxicology 335, 72–78 (2015).
Shirmanova, M. V. et al. Chemotherapy with cisplatin: insights into intracellular pH and metabolic landscape of cancer cells in vitro and in vivo. Sci. Rep. 7, 8911 (2017).
Loar, P. et al. Inhibition of glycolysis enhances cisplatin-induced apoptosis in ovarian cancer cells. Am. J. Obstet. Gynecol. 202, e371–e378 (2010).
Deliliers, G. L. et al. Effect of inositol hexaphosphate (IP(6)) on human normal and leukaemic haematopoietic cells. Br. J. Haematol. 117, 577–587 (2002).
Shu, Q. et al. Direct orthotopic transplantation of fresh surgical specimen preserves CD133+ tumor cells in clinically relevant mouse models of medulloblastoma and glioma. Stem Cells 26, 1414–1424 (2008).
Badodi, S., Marino, S. & Guglielmi, L. Establishment and culture of patient-derived primary medulloblastoma cell lines. Methods Mol. Biol. 1869, 23–36 (2019).
Conti, L. et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 3, e283 (2005).
Di Veroli, G. Y. et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics 32, 2866–2868 (2016).
Badodi, S., Baruffaldi, F., Ganassi, M., Battini, R. & Molinari, S. Phosphorylation-dependent degradation of MEF2C contributes to regulate G2/M transition. Cell Cycle 14, 1517–1528 (2015).
Dibenedetto, S. et al. Enhanced energetic state and protection from oxidative stress in human myoblasts overexpressing BMI1. Stem Cell Rep. 9, 528–542 (2017).
Hijazi, M., Smith, R., Rajeeve, V., Bessant, C. & Cutillas, P. R. Reconstructing kinase network topologies from phosphoproteomics data reveals cancer-associated rewiring. Nat. Biotechnol. 38, 493–502 (2020).
Wilkes, E. H., Terfve, C., Gribben, J. G., Saez-Rodriguez, J. & Cutillas, P. R. Empirical inference of circuitry and plasticity in a kinase signaling network. Proc. Natl Acad. Sci. USA 112, 7719–7724 (2015).
Cutillas, P. R. Targeted in-depth quantification of signaling using label-free mass spectrometry. Methods Enzymol. 585, 245–268 (2017).
Casado, P. et al. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci. Signal 6, rs6 (2013).
Dinkel, H. et al. Phospho.ELM: a database of phosphorylation sites–update 2011. Nucleic Acids Res. 39, D261–D267 (2011).
Hornbeck, P. V. et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015).
Yang, C. Y. et al. PhosphoPOINT: a comprehensive human kinase interactome and phospho-protein database. Bioinformatics 24, i14–i20 (2008).
Feber, A. et al. Using high-density DNA methylation arrays to profile copy number alterations. Genome Biol. 15, R30 (2014).
Maksimovic, J., Gordon, L. & Oshlack, A. SWAN: Subset-quantile within array normalization for illumina infinium HumanMethylation450 BeadChips. Genome Biol. 13, R44 (2012).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Tarazona, S. et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 43, e140 (2015).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Ramirez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 (2013).
Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Wu, G. & Haw, R. Functional interaction network construction and analysis for disease discovery. Methods Mol. Biol. 1558, 235–253 (2017).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Jones, K. M. et al. CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells 33, 196–210 (2015).
Whittaker, D. E. et al. The chromatin remodeling factor CHD7 controls cerebellar development by regulating reelin expression. J. Clin. Investig. 127, 874–887 (2017).
Yadirgi, G. et al. Conditional activation of Bmi1 expression regulates self-renewal, apoptosis, and differentiation of neural stem/progenitor cells in vitro and in vivo. Stem Cells 29, 700–712 (2011).
Acknowledgements
We are grateful to the BSU staff for help in the daily care of our mouse colony and to the Blizard Core Facilities (Imaging and FACS) for technical advice. Access to medulloblastoma tissue was obtained through the CCLG tissue bank (project number 2015 BS 05). This work is supported by grants of the Medical Research Council UK (MR/N000528/1) to S.M. and M.A.B. and a Brain Tumour Research Centre of Excellence award to S.M. Y.M.L. is funded by a Cancer Research UK Accelerator grant Cl 15121A 20256. Funding from Children with Cancer UK (15/188), Action Medical Research and The Brain Tumour Charity (GN2181) to C.D.B., S.C.C. and A.P. are also acknowledged. A.P. was funded by an NIHR Research Professorship (NIHR-RP-R2-12-019). S.Br. is partly supported by the Department of Health’s NIHR Biomedical Research Centre’s funding scheme to University College London Hospitals.
Author information
Authors and Affiliations
Contributions
S.Ba. and S.M. conceived the project, S.Ba., M.A.B. and S.M. designed the experiments, S.Ba. performed and analysed all the experiments, X.Z. and J.W. contributed to the animal experiments, M.V.N.C. co-designed the metabolic assays, N.P. analysed ChIP-Seq data, G.R. analysed DNA methylation and RNA-Seq data, Y.M.L. and S.Br. performed image analysis to quantify IHC results, G.M. and S.M.P. provided human foetal neural stem cells, C.D.B., S.C.C. and A.P. conducted ex vivo MRS experiments and S.Ba. and S.M. wrote the paper with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks Erik Berg Schmidt and the other anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Badodi, S., Pomella, N., Zhang, X. et al. Inositol treatment inhibits medulloblastoma through suppression of epigenetic-driven metabolic adaptation. Nat Commun 12, 2148 (2021). https://doi.org/10.1038/s41467-021-22379-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-021-22379-7
This article is cited by
-
13-Cis Retinoic Acid Induces Neuronal Differentiation in Daoy (Medulloblastoma) Cells Through Epigenetic Regulation of Topoisomerase IIβ
Applied Biochemistry and Biotechnology (2023)
-
Group-specific cellular metabolism in Medulloblastoma
Journal of Translational Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.